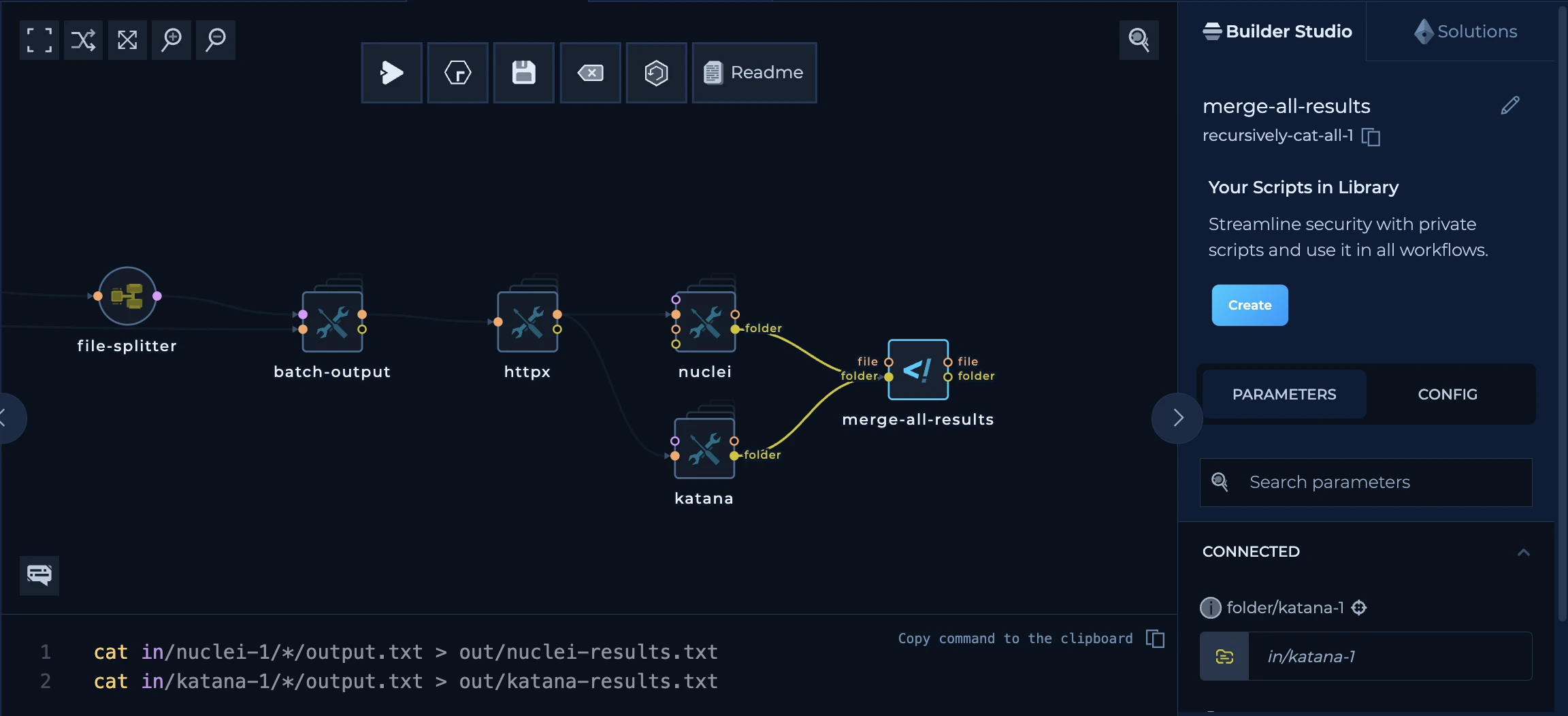
- Using Single Splitter Node
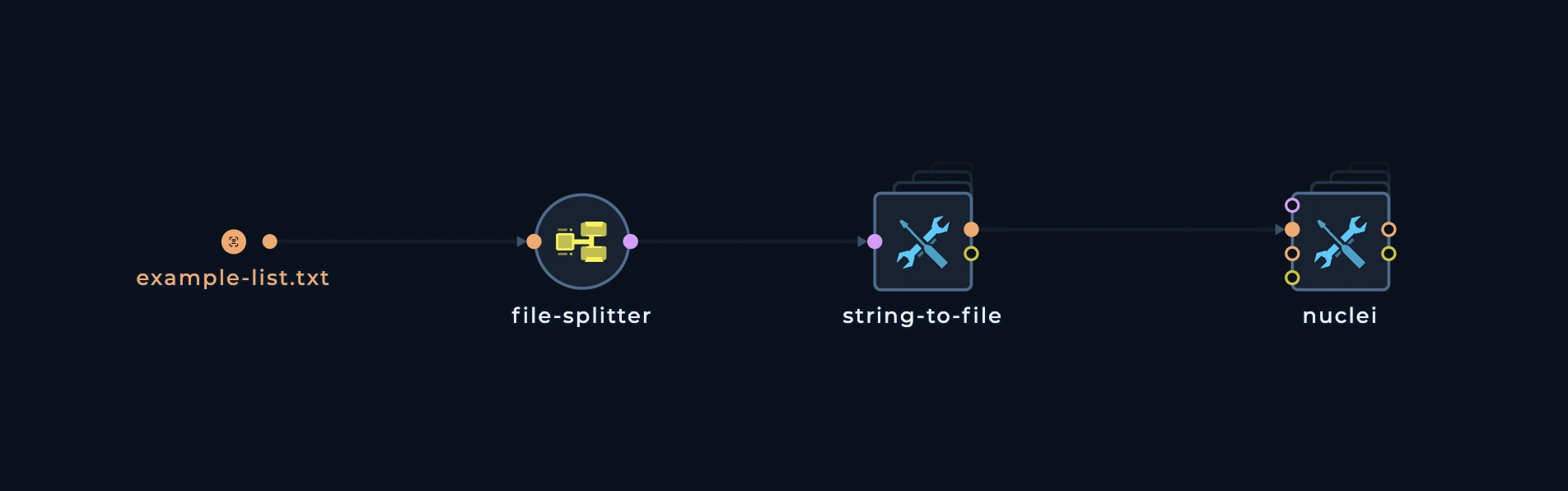
- Using batch output pattern
Using Single Splitter Node
Splitter node is a node that splits the input data into multiple strings. Let’s say you have a file calledto-distribute.txt. The file contains the following data:

When the distribution of the nodes is happening, platform will provide a visualization showing that multiple nodes are running in parallel
string-to-file which is a very basic one, and it is used to convert the string back to the file and help other tools continue the distribution. In our case, both string-to-file and nuclei will execute three times for each of domains described in the to-distribute.txt file.
Let’s say we want to stop nuclei results and merge everything, we can do it by connecting the recursively-cat-all (or any script) to the nuclei output and then connect the custom-script to the folder output.

in/nuclei-1/1/output.txt, in/nuclei-1/2/output.txt, in/nuclei-1/3/output.txt and merge them into one file out/output.txt.
Take a look at
recursively-cat-all script, you can see that visualization showing the distribution is gone, this means that splitter chain is stopped and that this script got all results in the file and folder structure mentioned above.Using batch output pattern
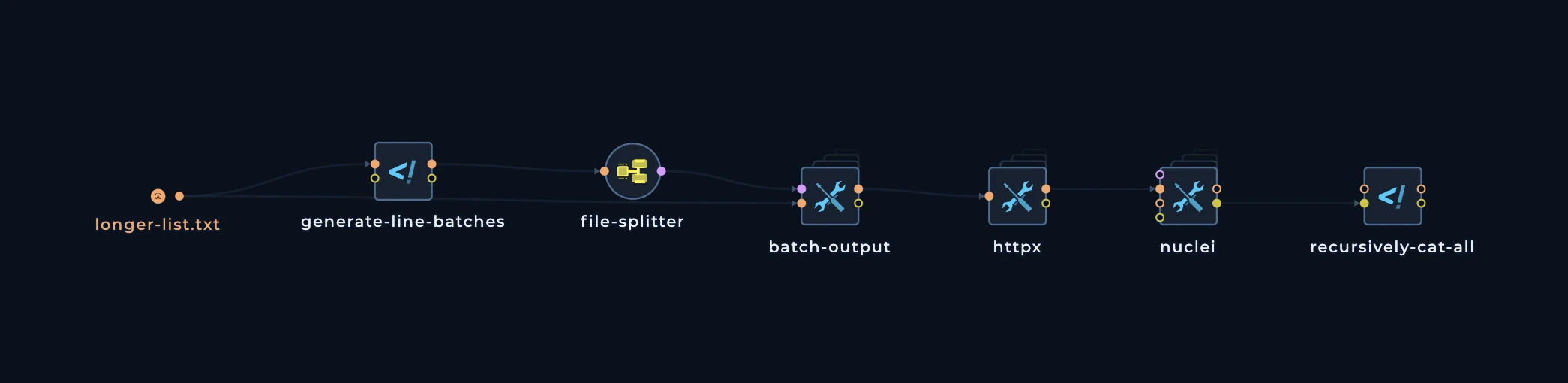
Now, let’s take a look at more complext pattern. This pattern is used when you have a lot of data to distribute and you want to distribute it in a more efficient way. This pattern is almost always tailored to tools which havefile inputs and you want to distribute the inputs in batches.
Let’s use a bigger file as example:
9 domains. We want to distribute these domains to multiple machines and to different tools. We can use a batch output pattern to split the data into multiple files and then connect these files to different nodes (tools).
Our fleet has 3 machines, and we want to distribute these domains to 3 machines. We can use a batch pattern to split the data into 3 files, each containing 3 domains. This pattern consists of 3 main nodes:
generate-line-batches
This node is used to calculate the number of lines in the input file length to be used forbatch-output node. It will generate a file with the number of lines in the input file.
batch-output
This node is used to split the input file into multiple files. It will split the input file into multiple files, each containing a specified number of lines. and of course, for this we will need to usefile-splitter
This is how the pattern looks like:
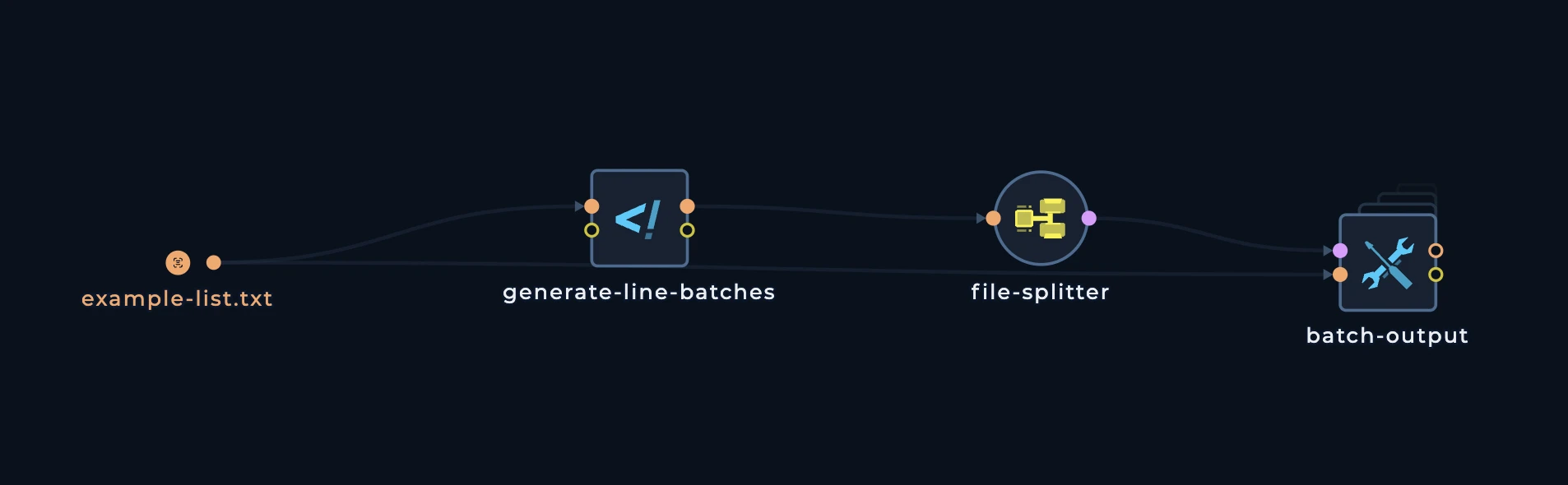
generate-line-batches script, we can see that it is calculating the number of lines in the input file and then it is creating a file with the number of lines in the input file.
BATCH_SIZE which is the number of lines in the input file that will be in each batch.
Notice also how the initial list is being passed to the batch-output node. So even if you don’t understand the script, you will be able to understand that this script is getting:
- Entire list of domains
- Lines from-to which will be used for each batch calculated by
generate-line-batchesscript
nuclei tool to scan these domains.
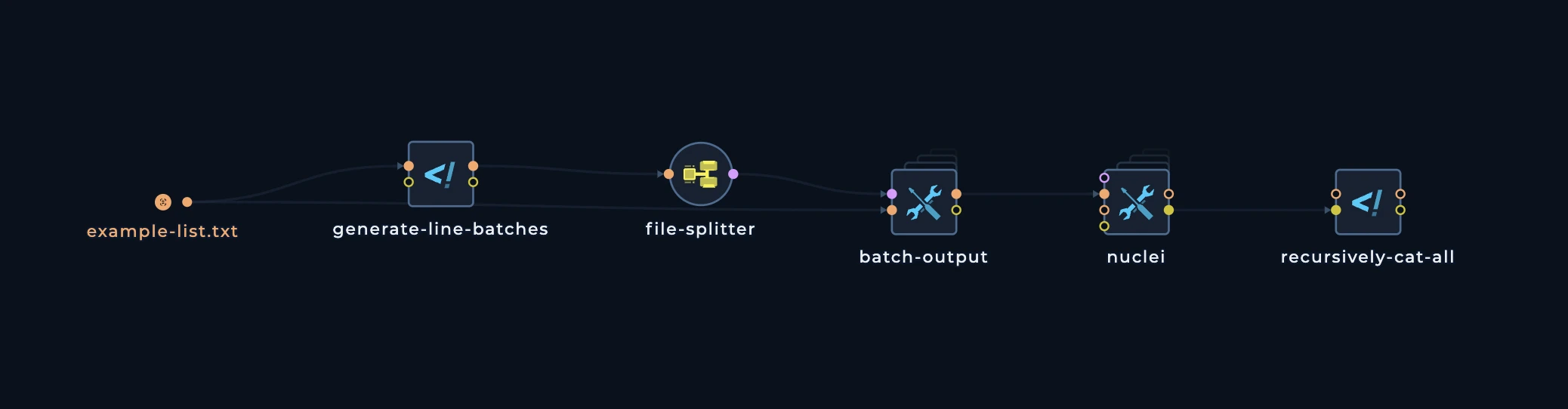
Continuing the distribution
Let’s take a list of much more subdomains, for example 10000. And we want to scan for web servers, do nuclei scan, and then merge all of the results. What we need to do:- Set the
BATCH_SIZEto100in thegenerate-line-batchesscript. In this case, we will have100domains in each batch, and thehttpxandnucleiwill each execute100times, which is less than current splitter limit. - Connect the
httpxto thebatch-outputoutput, and then connect thenucleito thebatch-outputoutput. - Connect the
recursively-cat-allscript to thenucleioutput, and then connect thecustom-scriptto thefolderoutput.
httpx and then create two separate files in recursively-cat-all script, one for nuclei and one for katana.
When we connect katana to httpx it will also execute 100 times. Just like nuclei will.
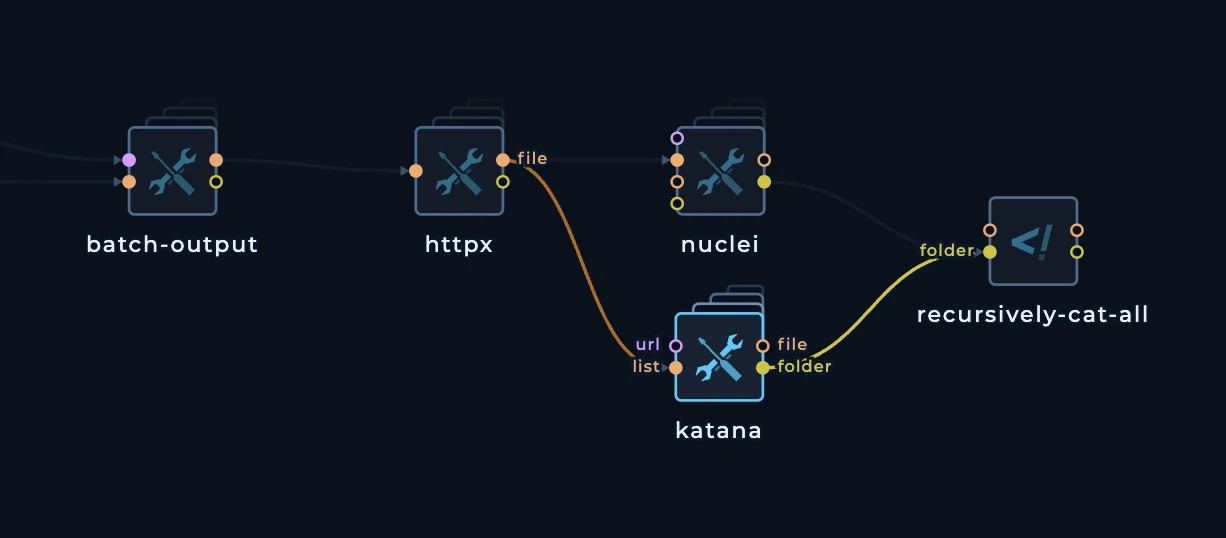
recursively-cat-all script to merge-all-results and change the script so we can have two files:
nuclei-results.txtkatana-results.txt
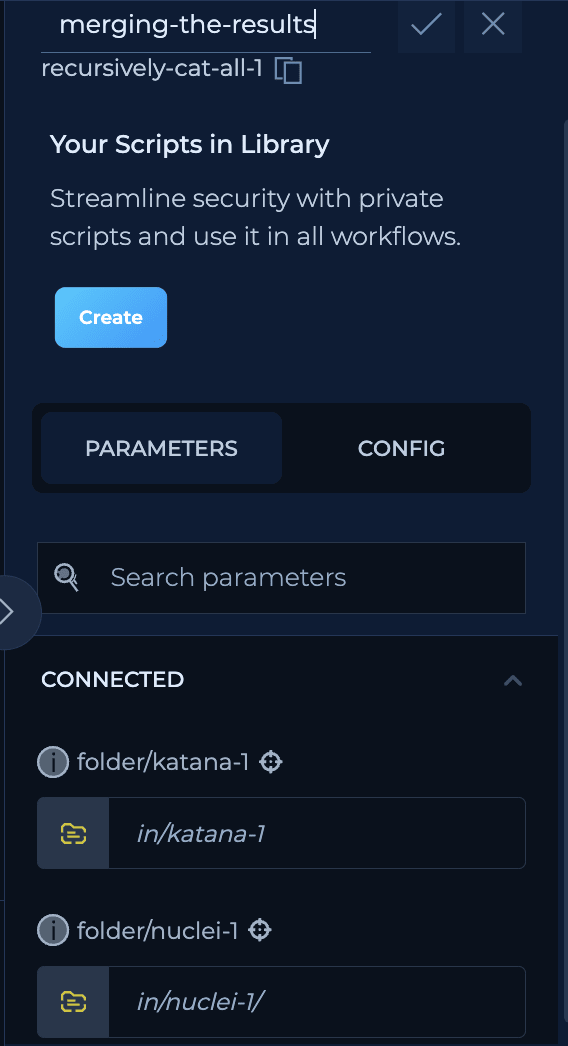 We have here two folders with this structure:
We have here two folders with this structure:
merge-all-results script we will have: